INTRODUCTION
The SYNTHIA™ Retrosynthesis software Shared Path Library allows for retrosynthesis analysis of up to 20 structurally related target molecules simultaneously, prioritizing pathways that share common intermediates and reactions among them. These analyses utilize proprietary algorithms that explore and rank reactions generated from an expert-coded database of chemistry rules.
The database of chemistry rules includes transformations commonly found in organic chemistry textbooks and literature. These rules have been hand-coded by PhD-level chemists and can account for incompatible functional groups in your reactions, as well as recognize functional groups that require protection.
This guide offers a step-by-step approach to setting up a Shared Path Library analysis using the SYNTHIA™ Retrosynthesis software and explains how the results are presented. A preloaded example of a Shared Path Library analysis sis is available in your account for your reference.
When you first use the software, we recommend experimenting with the same molecules to observe how changes in analysis settings impact your results.
The library used throughout this guide consists of three molecules selected from a series of analogues synthesized for a SAR study of SMURF1 ligase inhibitors. The SMILES strings are as follows:
COc1ccc(-c2onc(C(=O)Nc3c(C)n(C)n(-c4ccccc4F)c3=O)c2C)cc1.Cc1c(C(=O)Nc2c(C)n(C)n(-c3ccccc3F)c2=O)noc1-c1ccc(C2CC2)cc1Cl.Cc1c(C(=O)Nc2c(C)n(C)n(-c3ccccc3F)c2=O)noc1-c1ccc(Cl)cc1F
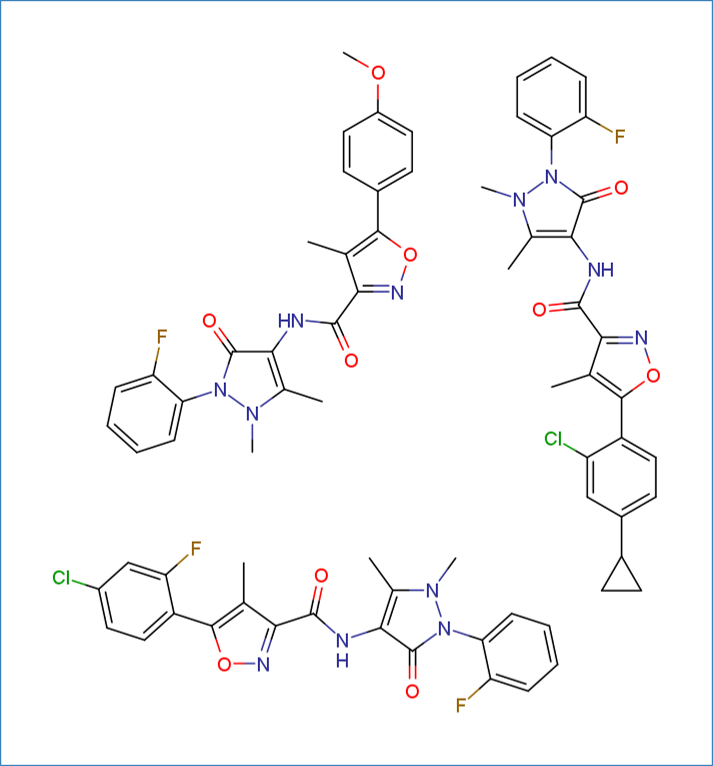
.png)

