INTRODUCTION
SYNTHIA™ Retrosynthesis software Retrosynthetic Analyses are based on our proprietary algorithms, which explore, and rank reactions generated from an expert-coded database of chemistry rules.
Retrosynthesis analyses completed with any of the Saved Configurations should return optimized results consistent with the selected Configuration. Yet, there may be times when getting desired results requires to dive deeper into the parameters that guide SYNTHIA™ Retrosynthesis software when exploring chemistry and ranking found pathways.
Automatic, Step-by-Step and Shared Path Library analyses can all be customized by modifying the parameters that direct the exploration of possible chemistry, determine which compounds are considered starting materials and rank which pathways are going to be returned in your results. This guide is designed to help you understand how to modify these parameters to get the results that are best fitted for your needs.
When you begin to use the software, we recommend that you experiment with the same molecule, so that you can see how changes in analysis settings affect your results.
The molecule used throughout this guide is sitagliptin, SMILES string:
N[C@@H](CC(=O)N1CCn2c(C1)nnc2C(F)(F)F)Cc1cc(F)c(F)cc1F
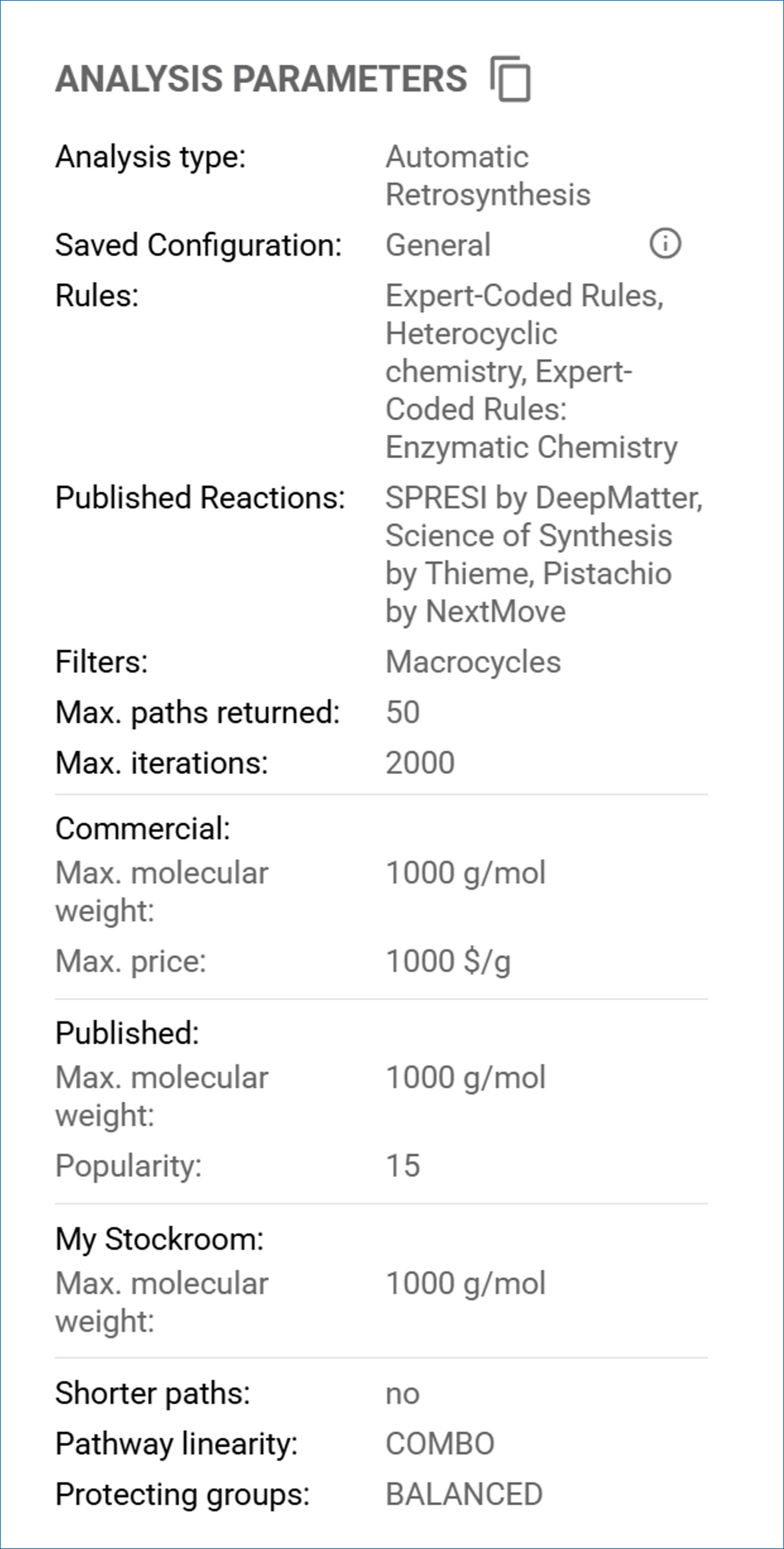
Note: To see how to setup a Retrosynthesis analysis with SYNTHIA™ Retrosynthesis software, you may refer to the following guides:
>> Automatic Retrosynthesis Analysis
>> Step-by-Step Retrosynthesis Analysis
>> Shared Path Library Analysis
.png)

